Attentional-blink tutorial (advanced)
- Difficulty
- The goal
- Step 1: Download and start OpenSesame
- Step 2: Choose template, font, and colors
- Step 3: Implement counterbalancing
- Step 4: Define experimental variables that are varied between blocks
- Step 5: Create instructions
- Step 6: Modify feedback
- Step 7: Define experimental variables that are varied within a block
- Step 8: Define trial sequence
- Step 9: Create RSVP stream (prepare phase)
- Step 10: Execute RSVP stream (run phase)
- Step 11: Create fixation point
- Step 12: Define response collection
- Step 13: Specify number and length of blocks
- Step 14: Run experiment!
- Extra 1: Check timing (and learn some NumPy)
- Extra 2: Add assertions to check your experiment
- Extra 3: Use PsychoPy directly
- References
Difficulty
This tutorial assumes a basic knowledge of OpenSesame, experimental design, and Python. An introductory OpenSesame tutorial can be found here:
Links to introductory Python tutorials can be found here:
The goal
In this tutorial, we will implement an attentional-blink paradigm, as introduced by Raymond, Shapiro, and Arnell (1992). We will re-create experiment 2 from Raymond et al. almost exactly, with only a few minor modifications. In this experiment, the participant sees a stream of letters, typically called an RSVP stream (for Rapid Serial Visual Presentation). There are two conditions. In the experimental condition, the participant's task is twofold:
- Report the identity of the white letter (all other letters were black).
- Indicate whether an 'X' was present.
In the control condition, the participant's task is only to ...
- Indicate whether an 'X' was present.
The white letter is called the T1 (or 'target'). The 'X' is called the T2 (or 'probe'). The typical finding is that the T2 is often missed when it is presented 200 - 500 ms after T1, but only when T1 needs to be reported. This phenomenon is called the attentional blink, because it is as though your mind's eye briefly blinks after seeing T1. But surprisingly, T2 is usually not missed when it follows T1 immediately. This is called lag-1 sparing. The results of Raymond et al. (1992) looked like this:
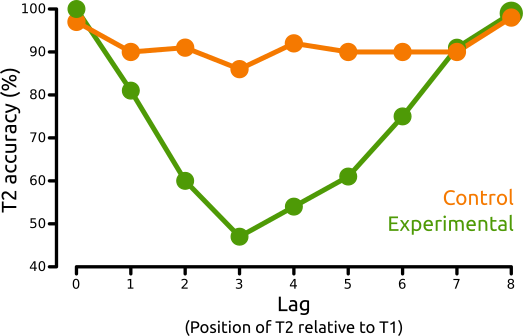
Figure 1. T2 accuracy as a function of the serial position of T2 relative to T1 ('lag'). A lag of 0 means that T1 and T2 where identical (i.e. a white 'X'). Adapted from Raymond et al. (1992).
Step 1: Download and start OpenSesame
OpenSesame is available for Windows, Linux, Mac OS (experimental), and Android (runtime only). This tutorial is written for OpenSesame 3.0.X. You can download OpenSesame from here:
When you start OpenSesame, you will be given a choice of template experiments, and a list of recently opened experiments (Figure 2).
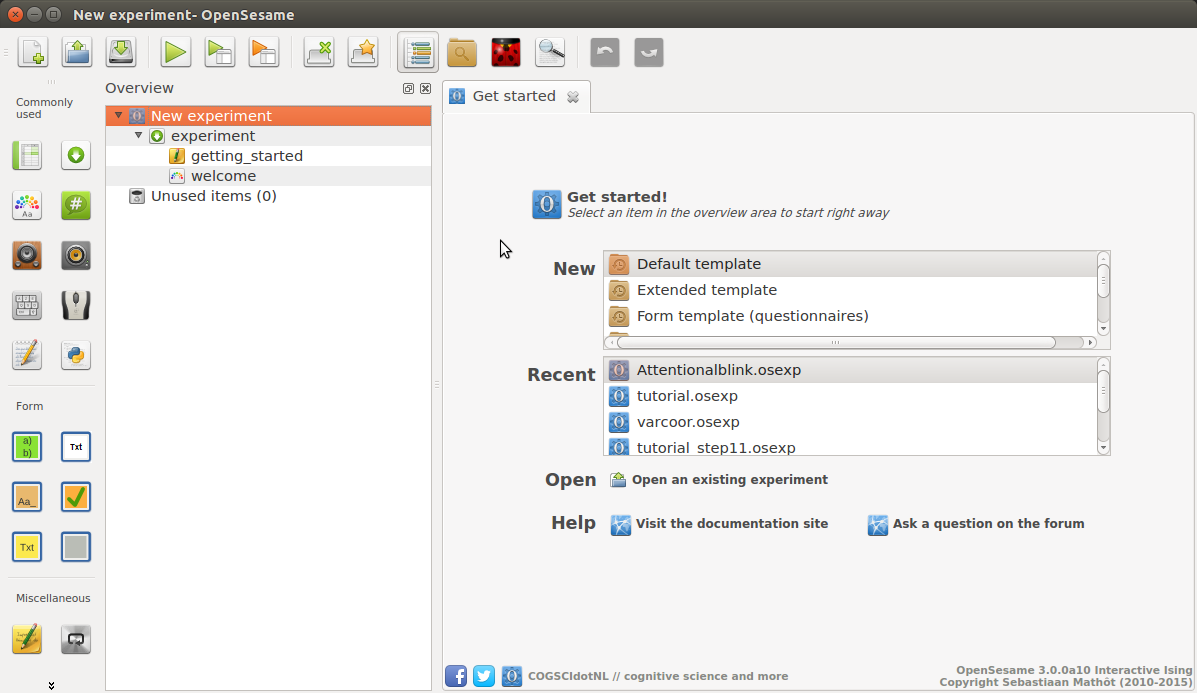
Figure 2. The OpenSesame window on start-up.
Step 2: Choose template, font, and colors
The 'Extended template' provides the basic structure of a typical trial-based experiment with a practice and experimental phase. Because our experiment fits this template very well, we're going to use it. Therefore, double-click on 'Extended template' to open it.
In the 'General tab' that now appears, you can specify the general properties of your experiment. For this experiment, we want to use black letters on a gray background. Also, the default font size of 18 is a bit small, so change that to 32. Finally, it's good practice to give your experiment an informative name and description. Your 'General tab' now looks as in Figure 3.
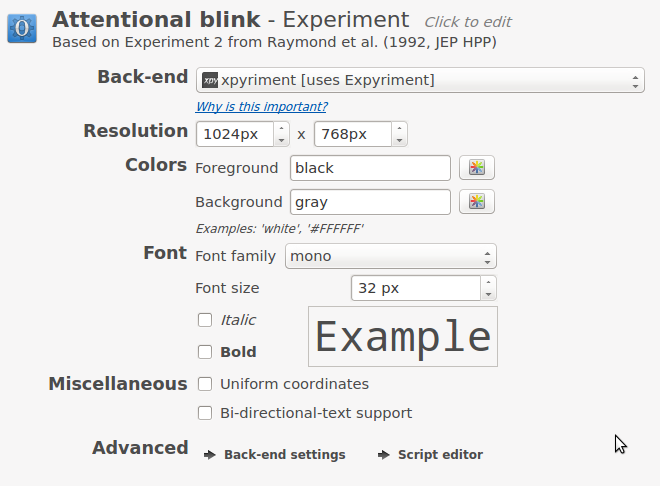
Figure 3. The General tab is where you define the general properties of your experiment.
Step 3: Implement counterbalancing
In Raymond et al. (1992), the experimental and control conditions were mixed between blocks: Participants first did a full block in one condition, and then a full block in the other condition. Condition order was counterbalanced, so that half the participants started with the experimental condition, and the other half started with the control condition.
Let's start with the counterbalancing part, and use the participant number to decide which condition is tested first. We need to do this as the very first thing of the experiment, and we need to use some Python scripting to do it.
Therefore, drag an inline_script from the item toolbar onto the very top of the experiment. Change the name of the new item to counterbalance. In the Prepare phase of the counterbalance item, enter the following script:
if var.subject_parity == 'even':
var.condition1 = 'experimental'
var.condition2 = 'control'
else:
var.condition1 = 'control'
var.condition2 = 'experimental'
Ok, let's take a moment to understand what's going on here.
The first thing to know is that experimental variables are properties of the var object. Experimental variables are variables that you have defined yourself, for example in a loop item, as well as built-in variables. One such built-in experimental variable is subject_parity, which is automatically set to 'even' when the experiment is launched with an even subject number (0, 2, 4, etc.), and to 'odd' when the subject number is odd (1, 3, 5, etc.).
We further create two new experimental variables condition1 and condition2. By setting these as properties of var, we make them available elsewhere in OpenSesame, outside of inline_script items. So this line:
var.condition1 = 'experimental'
... creates an experimental variable with the name condition1, and gives it the value 'experimental'. In step 4, we will use this variable to determine which condition is tested first.
In other words, this script says the following:
- All even-numbered subjects start with the experimental condition.
- All odd-subjects start with the control condition.
Step 4: Define experimental variables that are varied between blocks
As mentioned above, conditions are varied between blocks. To understand how this works in OpenSesame, it's best to start at the bottom (see Figure 4), with ...
- the trial_sequence, which corresponds (as you might expect) to a single trial. One level above ...
- the block_loop corresponds to a single block of trials. Therefore, this is where you would define experimental variables that are varied within a block. One level above ...
- the block_sequence corresponds to a single block of trials plus the events that happen before and after every block, such as post-block feedback on accuracy, and pre-block instructions. One level above ...
- the practice_loop and experimental_loop correspond to multiple blocks of trials during respectively the practice and non-practice (experimental) phase. Therefore, this is where you would define experimental variables that are varied between blocks.
In other words, we need to define our between-block manipulations near the top of the experimental hierarchy, in the practice_loop and experimental_loop.
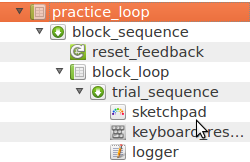
Figure 4. A fragment of the experimental structure as shown in the overview area.
Click on practice_loop to open the item. Right now, there is only one variable, practice, which has the value 'yes' during one cycle (i.e. one block).
Let's get to work! Add a variable called condition, change the number of cycles to 2, and change the order to 'sequential'.
Now use the previously created variables condition1 and condition2 to determine which condition is executed first, and which second (see Figure 5). To indicate that something is the name of a variable, and not a literal value, put square brackets around the variable name: '[my_variable]'
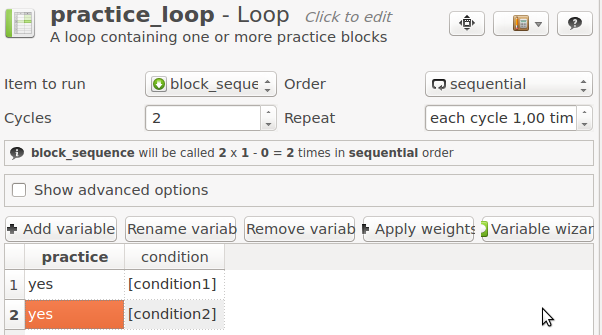
Figure 5. The practice_loop item after Step 4.
Do the same thing for experimental_loop, except that the variable practice has the value 'no'. (The practice variable doesn't have a real function. It only allows you to easily filter out all practice trials during data analysis.)
Step 5: Create instructions
Because the task differs between blocks, we need to show an instruction screen before each block. The block_sequence is the place to do this, because, as explained above, it corresponds to a single block of trials plus the events that occur before and after every block.
There are various items that we could use for an instruction screen, but we will use the sketchpad. Insert two new sketchpads at the top of block_sequence by dragging them from the item toolbar. Rename the sketchpads to instructions_experimental and instructions_control. Click on both items to add some instructional text, such as shown in Figure 6.
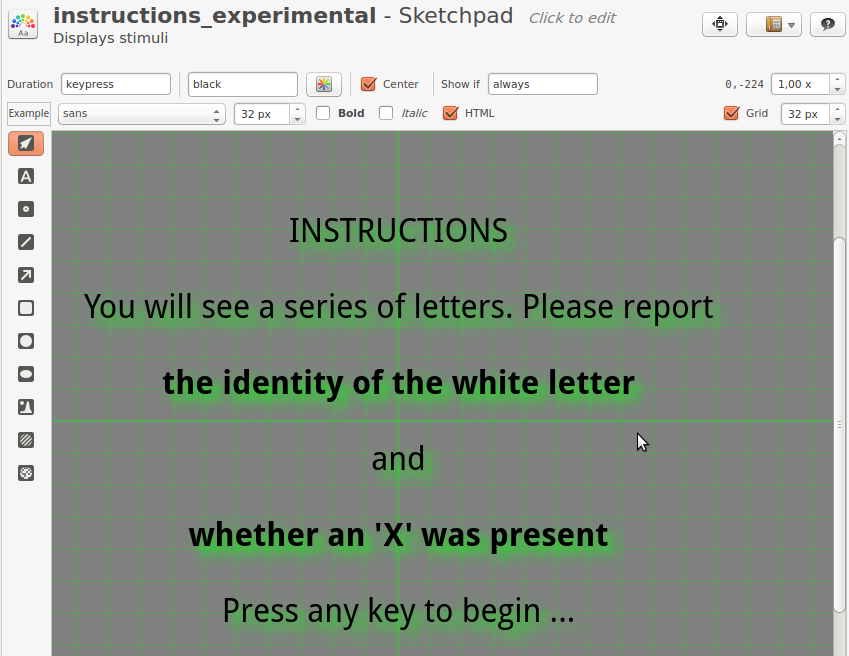
Figure 6. An example of instructional text in the instructions_experimental item.
Right now both instruction screens are shown before every block, which is not what we want. Instead, we want to show only the instructions_experimental item in the experimental condition, and only the instructions_control item in the control condition. We can do this with conditional ('run if') statements.
Click on block_sequence to open it. You will see a list of item names, just as in the overview area, except that each item has the text 'always' next to it. These are run-if statements, and they determine the conditions under which an item is executed. Double click on the run-if statement next to instructions_experimental and add the following text:
[condition] = experimental
This means that instructions_experimental will only be executed when the variable condition has the value 'experimental.' Analogously, change the run-if statement for instructions_control to:
[condition] = control
Your block_sequence should now look as in Figure 7.
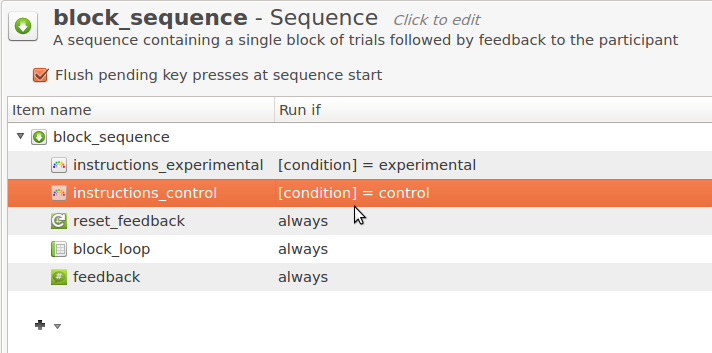
Figure 7. The block_sequence item at the end of Step 5.
Step 6: Modify feedback
Open feedback. By default, in the Extended Template, the participant receives feedback on speed (avg_rt) and accuracy (acc) after each experimental block. However, our experiment doesn't require speeded responses, and we should therefore only provide feedback on accuracy. Modify the feedback item to look something like Figure 8.
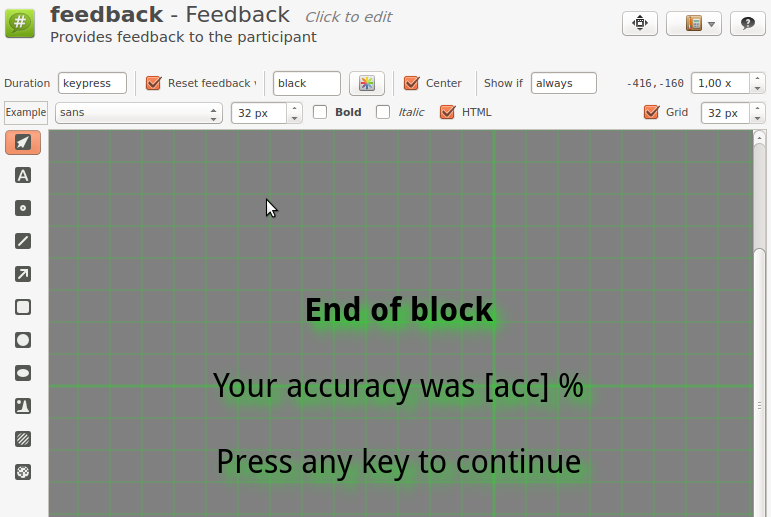
Figure 8. The block_sequence item at the end of Step 6.
Links
Step 7: Define experimental variables that are varied within a block
Raymond et al. (1992) vary the position of T2 relative to T1 from 0 to 8, where 0 means that one letter is both T1 and T2 (i.e. a white 'X'). They also have trials in which there is no T2. This is all varied within a block. There are various ways to code this, but the easiest way is to use two variables:
lagindicates the position of T2 relative to T1. It has a value of 0 - 8, or no value if there is no T2.T2_presentis 'y' for trials on which there is a T2 and 'n' for trials on which there is no T2. Of course, this is redundant, becauseT2_presentis 'y' on all trials on whichlaghas a value. But it's convenient to defineT2_present, because we can use it later on to specify the correct T2 response.
Click on block_loop and create a variable table as shown in Figure 9.
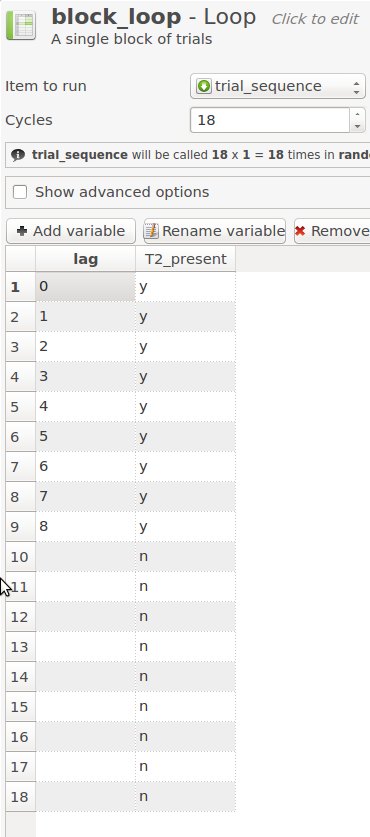
Figure 9. The block_loop item after Step 7.
Step 8: Define trial sequence
We will use an inline_script item to do most of the heavy lifting, and therefore our trial_sequence is quite simple. It consists of:
- A sketchpad (called fixation) to show a fixation dot.
- An inline_script (called RSVP) item that implements the RSVP stream.
- A sketchpad (called ask_T1) that asks the participant to report T1.
- A keyboard_response (called response_T1) that collects the T1 report.
- A sketchpad (called ask_T2) that asks the participant to report T2.
- A keyboard_response (called response_T2) that collects the T2 report.
- A logger (called logger) that writes all the data to a log file.
Drag all the required items from the item toolbar into trial_sequence, re-order them if necessary, and give them informative names. Also, use run-if statements to collect a T1 response only in the experimental condition. Your trial sequence should now look like Figure 10.
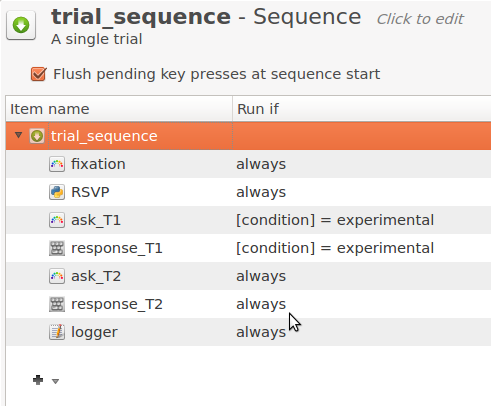
Figure 10. The trial_sequence item after Step 8.
Step 9: Create RSVP stream (prepare phase)
Now we're getting to the fun-but-tricky part: implementing the RSVP stream. Click on RSVP to open the item. You see two tabs: Prepare and Run. The golden rule is to add all code related to stimulus preparation to the Prepare tab, and all code related to stimulus presentation to the Run tab. Let's start with the preparatory stuff, so switch to the Prepare tab.
First, we need to import the Python modules that we plan to use:
import random
import string
Next, we need to define several variables that determine the details of the RSVP stream. We will make them properties of the var object, that is, turn them into experimental variables. This not necessary, but has the advantage that they will be automatically logged.
# The color of T1
var.T1_color = 'white'
# The presentation time of each stimulus
# (rounded up to nearest value compatible with refresh rate)
var.letter_dur = 10
# The inter-stimulus interval
# (rounded up to nearest value compatible with refresh rate)
var.isi = 70
Next, we are going to create the letter stream. Raymond et al. have a few rules:
- The number of letters that precede T1 is randomly selected between 7 and 15.
- The number of letters that follow T1 is always 8.
- Letters are randomly sampled without replacement from all uppercase letters except 'X' (which is used for T2).
Let's translate these rules to Python:
# The position of T1 is random between 7 and 15. Note that the first position is
# 0, so the position indicates the number of preceding stimuli.
var.T1_pos = random.randint(7, 15)
# The maximum lag, i.e. the number of letters that follow T1.
var.max_lag = 8
# The length of the stream is the position of T1 + the maximum lag + 1. We need
# to add 1, because we count starting at 0, so the length of a list is always
# 1 larger than its maximum index.
var.stream_len = var.T1_pos + var.max_lag + 1
# We take all uppercase letters, which have been predefined in the `string`
# module. Converting to a `list` creates a list of characters.
letters = list(string.ascii_uppercase)
# We remove 'X' from this list.
letters.remove('X')
# Randomly sample a `stream_len` number of letters
stim_list = random.sample(letters, var.stream_len)
Ok, stim_list now contains all letters that make up our RSVP stream on a given trial, except for the T2 (if present). Therefore, on T2-present trials, we need to replace the letter at the T2 position by an 'X'.
if var.T2_present == 'y':
var.T2_pos = var.T1_pos + var.lag
stim_list[var.T2_pos] = 'X'
We now have a variable called stim_list that specifies the letters in our RSVP stream. This is a list that might contain something like: ['M', 'F', 'O', 'P', 'S', 'R', 'Y', 'C', 'U', 'Z', 'G', 'A', 'T', 'E', 'H', 'J', 'V', 'N', 'B', 'K', 'X', 'Q']. Note that stim_list is not an experimental variable, i.e. it is not a property of the var object. This is because experimental variables cannot be lists: The var object would turn the list into a character string, and that's not what we want!
The next step is to create a list of canvas objects, each of which contains a single letter from stim_list. A canvas object corresponds to a static visual stimulus display, i.e. to one frame in our RSVP stream. You can create a canvas object using the canvas() function, which is one of OpenSesame's common
functions that you can call without needing to import anything.
# Create an empty list for the canvas objects.
letter_canvas_list = []
# Loop through all letters in `stim_list`. `enumerate()` is a convenient
# function that automatically returns (index, item) tuples. In our case, the
# index (`i`) reflects the position in the RSVP stream. This Python trick, in
# which you assign a single value to two variables, is called tuple unpacking.
for i, stim in enumerate(stim_list):
# Create a `canvas` object.
letter_canvas = canvas()
# If we are at the position of T1, we change the foreground color, because
# T1 is white, while the default color (specified in the General tab) is
# black.
if i == var.T1_pos:
letter_canvas.set_fgcolor(var.T1_color)
# Draw the letter!
letter_canvas.text(stim)
# And add the canvas to the list.
letter_canvas_list.append(letter_canvas)
We also need to create a blank canvas to show during the inter-stimulus interval:
blank_canvas = canvas()
Finally, we set the identity of T1 as an experimental variable, because it has been randomly determined in the script:
# Extract T1 from the list
var.T1 = stim_list[var.T1_pos]
Preparation done!
Links
Step 10: Execute RSVP stream (run phase)
Now, let's switch to the Run tab of the RSVP item. Here we add the code that is necessary to show all the canvas objects that we have created during the Prepare phase. And that's not so hard! All we need to do is:
- For each letter canvas in the letter-canvas list
- Show the letter canvas
- Wait for
letter_durmilliseconds - Show the blank canvas
- Wait for
isimilliseconds
This translates almost directly into Python:
for letter_canvas in letter_canvas_list:
letter_canvas.show()
clock.sleep(var.letter_dur)
blank_canvas.show()
clock.sleep(var.isi)
Done!
Step 11: Create fixation point
After all this coding, it's time to get back to something simpler: Defining the fixation point. Click on fixation in to open the item. Change the duration to 995. This value will be rounded up to the nearest value compatible with your monitors refresh rate, which is 1000 ms for most common refresh rates. Draw a fixation dot in the center, using the fixation-dot tool (the dot with the little hole in it).
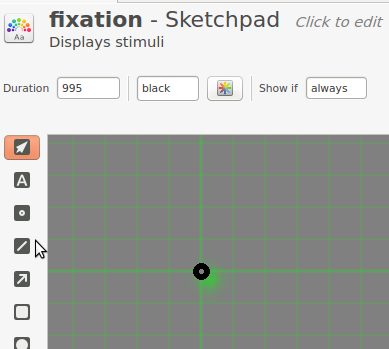
Figure 11. The fixation sketchpad after Step 11.
Step 12: Define response collection
We will collect responses as follows:
- Ask for T1
- Collect a response, which is a single key press that corresponds to T1. So if T1 was 'A', the participant should press the 'a' key.
- Ask for T2
- Collect a response, which is 'y' when T2 was present and 'n' when T2 was absent.
We will use the ask_T1 sketchpad to ask the participant for T1. Click on ask_T1 to open the item, and add a line of text, such as 'Please type the white letter'. Change the duration to 0. This 0 ms duration does not mean that the text is only shown for 0 ms, but that the experiment moves immediately to the next item, which is response_T1.
Open response_T1. The only thing that we have to do is define the correct response. To do this, we can use the T1 experimental variable that we have set while preparing the RSVP stream. Therefore, enter '[T1]' in the 'Correct response' field.
Open ask_T2, and add a line of text, such as 'Did you see an X? (y/n)'. Again, set the duration to 0, so that the experiment moves immediately to the next item, which is response_T2.
Open response_T2. Again, we need to define the correct response, this time using the variable T2_present, which we had defined in the block_loop. Therefore, add '[T2_present]' to the 'Correct response' field. It's also useful to restrict the allowed responses to 'y' and 'n', so that participants don't accidentally press the wrong key. You can do this by entering a semicolon-separated list of keys in the 'Allowed responses' field (i.e. 'y;n').
So how will the responses be logged? Each response item sets response, correct, and response_time variables. In addition, to distinguish responses set by different items, each response item sets these same variables followed by _[item name]. In other words, in this experiment the response variables of interests would be correct_T1_response and correct_T2_response.
Step 13: Specify number and length of blocks
You now have a fully working experiment, but one thing still needs to be done: Setting the length and number of blocks. We will use the following structure:
- 1 practice block of 9 trials in each conditon.
- 5 experimental blocks of 36 trials in each condition.
First, open block_loop. The 'Repeat' value is currently set to 1, which means that each trial is executed once, giving a block length of 18 trials. We want to specify the 'Repeat' value with a variable, so that we can have a different value for the practice and experimental blocks. To do this, we need to make a small modification to the script of block_loop. Click on the 'View' button in top-right of the tab (the middle of the three buttons), and select 'View script'. This will hide the graphical controls, and show the underlying OpenSesame script. Now change this line ...
set repeat "1"
... to ...
set repeat "[block_repeat]"
... and click 'Apply and close'. This means that the variable repeat is now defined in terms of another variable, block_repeat. OpenSesame will tell you that it doesn't know the length of the block anymore (see Figure 12), but that's ok: As long as the variable block_repeat is defined, things will work fine.
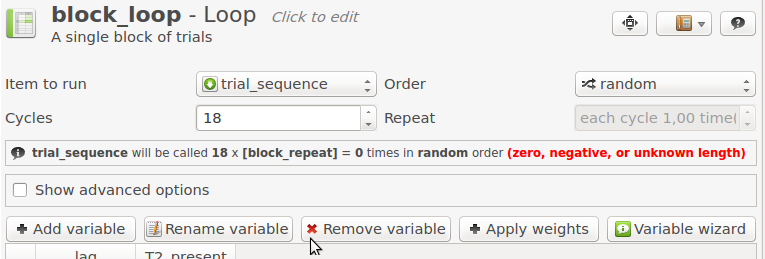
Figure 12. If the length of a loop is variably defined, OpenSesame notifies you of this.
Now open practice_loop. Add a variable block_repeat and give it the value 0.5. This means that 0.5 x 18 = 9 cycles of block_loop will be executed, just as we want.
Now open experimental_loop. Again, add a variable block_repeat and give it the value 2. This means that each block has a length of 2 x 18 = 36 trials. Also, change the number of cycles to 10, and arrange the loop table so that you first have five blocks of condition1, followed by five blocks of condition2 (see Figure 13).
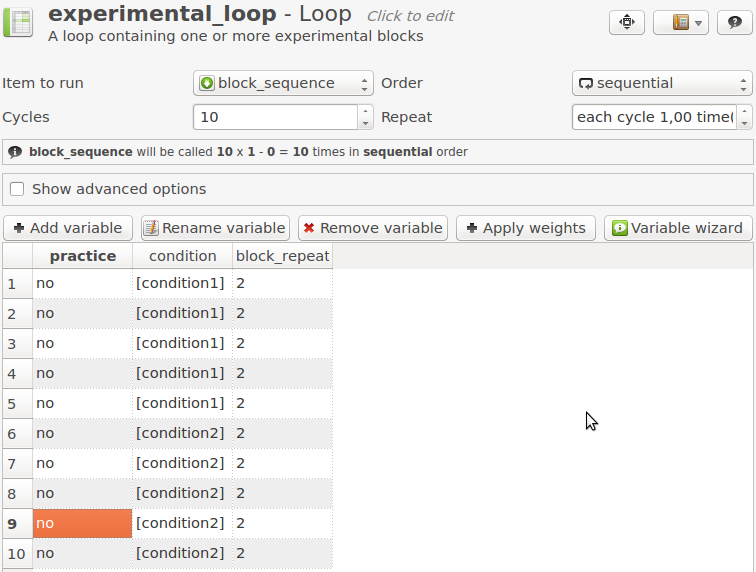
Figure 13. If the length of a loop is variably defined, OpenSesame notifies you of this.
Step 14: Run experiment!
That's it. You can now run the experiment!

Figure 14. Yes, you did!
Extra 1: Check timing (and learn some NumPy)
In time-critical experiments, you should always verify whether the timing is as intended. When using canvas objects, you can make use of the fact that the canvas.show() method returns the timestamp of the display onset. Therefore, as a first step, we maintain two lists: one to keep track of the letter-canvas onsets, and one to keep track of the blank-canvas onsets.
To do this, we need a small modification to the script in the Run tab of the RSVP item:
l_letter_time = []
l_blank_time = []
for letter_canvas in letter_canvas_list:
t1 = letter_canvas.show()
l_letter_time.append(t1)
clock.sleep(var.letter_dur)
t2 = blank_canvas.show()
l_blank_time.append(t2)
clock.sleep(var.isi)
We now have two lists with timestamps: l_letter_time and l_blank_time From these, we want to determine the average presentation duration of a letter, the average duration of a blank, and the standard deviation for both averages. But because lists are not great for these kinds of numerical computations, we are going to convert them to another kind of object: a numpy.array.
import numpy
a_letter_time = numpy.array(l_letter_time)
a_blank_time = numpy.array(l_blank_time)
Now we can easily create an array that contains the presentation duration for each letter:
a_letter_dur = a_blank_time - a_letter_time
This creates a new array, a_letter_dur, in which each item is the result of subtracting the corresponding item in a_letter_time from the corresponding item in a_blank_time. Schematically:
a_letter_dur -> [ 1, 1, 1 ]
=
a_blank_time -> [ 11, 21, 31 ]
-
a_letter_time -> [ 10, 20, 30 ]
Similarly, but slightly more complicated, we can create a new array, a_blank_dur, in which each item is the result of subtracting item i in a_blank_time from item i+1 in a_letter_time.
a_blank_dur = a_letter_time[1:] - a_blank_time[:-1]
Schematically:
a_blank_dur -> [ 9, 9 ]
=
a_letter_time[1:] -> [ 20, 30 ] # The leading 10 is stripped off
-
a_blank_time[:-1] -> [ 11, 21 ] # The trailing 31 is stripped off
The next step is to use the array.mean() and array.std() methods to get the averages and standard deviations of the durations in one go. To inspect these
values, we set them as experimental variables (i.e. as properties of the var object). That way they will be logged and visible in the variable inspector.
var.mean_letter_dur = a_letter_dur.mean()
var.std_letter_dur = a_letter_dur.std()
var.mean_blank_dur = a_blank_dur.mean()
var.std_blank_dur = a_blank_dur.std()
Done!
Extra 2: Add assertions to check your experiment
A Dutch proverb states that a mistake is in a small corner. (I suspect that according to the original proverb the mistake, rather than the corner, was small, but no matter.) Developing experiments, or any kind of software, without bugs is almost impossible. However, you can protect yourself from many bugs by building safeguards into your experiment.
For example, our experiment has two conditions, defined as 'experimental' and 'control'. But what if I accidentally misspelled 'experimental' as 'experimentel' in the experimental_loop? The experiment would still run, but it would no longer work as expected. Therefore, we want to make sure that condition is either 'experimental' or 'control', but nothing else. In computer-speak, we want to assert that this is the case. Let's take a look at how we can do this.
First, drag a new inline_script item to the start of the trial_sequence and rename it to assertions. Add the following line to the Run tab:
assert(var.condition in ['experimental', 'control'])
Let's dissect this line:
var.conditionrefers to the experimentalconditionvariable.in ['experimental', 'control']checks whether this variable matches any of the items in the list, i.e. whether it is 'experimental' or 'control'.assert()states that there has to be a match. If not, the experiment will crash (anAssertionErrorwill be raised).
In other words, whatever you pass to assert() has to be True, otherwise your experiment will crash. This useful for sanity checks.
Some more assertions:
assert(var.T2_present in ['y', 'n'])
assert(var.lag in ['']+range(0,9))
And a final one that is a bit more complicated. Can you figure out what it does?
assert((var.lag == '') != (var.T2_present == 'y'))
Links
- https://wiki.python.org/moin/UsingAssertionsEffectively
- Advice on protective programming in Axelrod (2014, doi:10.3389/fpsyg.2014.01435)
Extra 3: Use PsychoPy directly
OpenSesame is backend independent. This means that different libraries can be used for controlling the display, sound, response collection, etc. You can select the backend in the General tab.
So far, we have used OpenSesame's own canvas object, which automatically maps onto the correct functions of the selected backend. Therefore, you don't have to bother with or know about the details of each backend. However, you can also directly use the functions offered by a specific backend, such as PsychoPy. This is especially useful if you want to use functionality that is not available in OpenSesame's own modules.
First, to use PsychoPy, you need to switch to the psycho backend, which you can do in the 'General properties' tab of your experiment . Now, when you start the experiment, OpenSesame will automatically initialize PsychoPy, and the psychopy.visual.Window object will be available as win in inline_scripts.
Now let's see how we can implement our RSVP stream in PsychoPy. (The script below replaces the part in the Prepare phase of RSVP in which we created letter_canvas_list.)
from psychopy import visual
textstim_list = []
for i, stim in enumerate(stim_list):
if i == var.T1_pos:
color = 'white'
else:
color = 'black'
# All stimuli require an psychopy.visual.Window object to be passed as first
# argument. In OpenSesame, this object is available as `win`.
textstim = visual.TextStim(win, text=stim, color=color)
textstim_list.append(textstim)
The main difference with our previous script is that we don't draw text on a canvas object. Instead, the text is an object by itself (a TextStim), and it has its own draw() method to draw it to the screen.
Of course, we also need to update the Run phase of the RSVP stream, which now looks like this:
for textstim in textstim_list:
textstim.draw()
win.flip()
clock.sleep(var.letter_dur)
win.flip()
clock.sleep(var.isi)
The main difference here is that we need to call several methods to show our stimuli, instead of only canvas.show(). First, we need to call the draw() method on all stimuli that we want to show: textstim.draw() Next, we need to call win.flip() to refresh the display so that the stimuli actually become visible. If we call win.flip() without any preceding calls to draw(), as we do before the inter-stimulus-interval, it has the effect of clearing the display.
That's it!
References
Axelrod, V. (2014). Minimizing bugs in cognitive neuroscience programming. Frontiers in Psychology: Perception Science, 5, 1435. doi:10.3389/fpsyg.2014.01435
Mathôt, S., Schreij, D., & Theeuwes, J. (2012). OpenSesame: An open-source, graphical experiment builder for the social sciences. Behavior Research Methods, 44(2), 314–324. doi:10.3758/s13428-011-0168-7
Peirce, J. W. (2007). PsychoPy: Psychophysics software in Python. Journal of Neuroscience Methods, 162(1-2), 8–13. doi:10.1016/j.jneumeth.2006.11.017
Raymond, J. E., Shapiro, K. L., & Arnell, K. M. (1992). Temporary suppression of visual processing in an RSVP task: An attentional blink? Journal of Experimental Psychology: Human Perception and Performance, 18(3), 849–860. doi:10.1037/0096-1523.18.3.849






